Museum specimens serve as windows into the past through which we can observe how species have coped (or not) with environmental change. One way of measuring organismal response is through interrogation of the epigenome, however, the vast majority of museum specimens were not preserved with the notion of later extracting epigenomic data. Thus, the ability to track temporal epigenomic changes appears limited …. or is it?
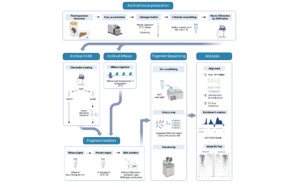
My team has opened a pathway to recovering temporal epigenetic change by linking two disparate fields – museum curation and chromatin biology – through their common use of the preservative formaldehyde. Our approach views formaldehyde fixation as a perk rather than an impediment to molecular analyses. By crosslinking nucleosomes in-place, formaldehyde fixation enables a window into historical chromatin accessibility.
Our approach – along with demonstration in specimens up to 117 years old – is currently available as a preprint. Our unique methodological and analytical approach enables the first ever detailed view of the epigenetic past and demonstrates a new role for museum collections in understanding chromatin architecture dynamics over the last century.
For an additional perspective on the potential for impact of a temporal epigenomic perspective, see our 2020 paper in TREE.